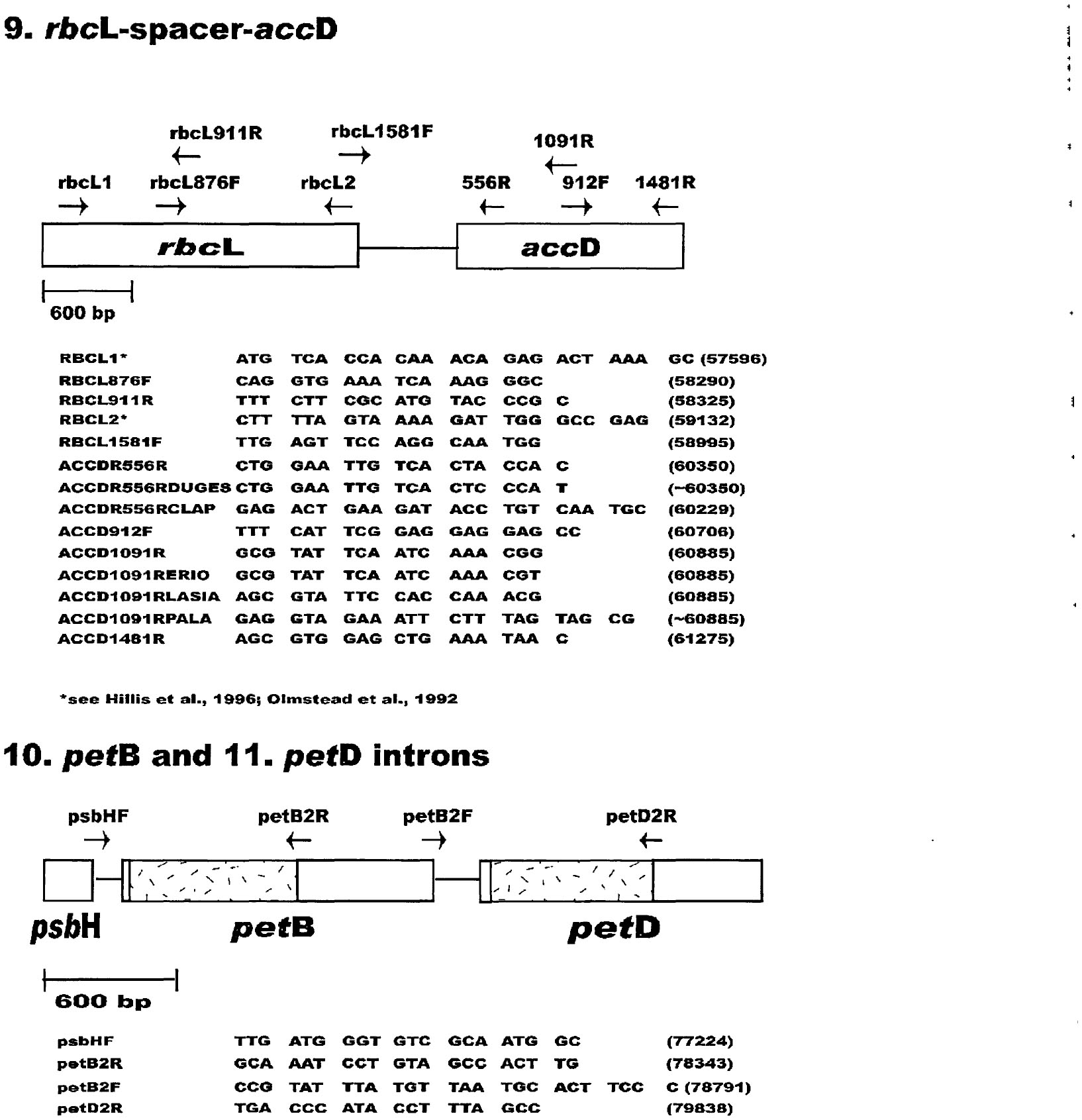
![PDF] Refinements for the amplification and sequencing of red algal DNA barcode and RedToL phylogenetic markers: a summary of current primers, profiles and strategies | Semantic Scholar PDF] Refinements for the amplification and sequencing of red algal DNA barcode and RedToL phylogenetic markers: a summary of current primers, profiles and strategies | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/4d9f21f79639203014634cba18dbb4f8ad3555f9/8-Figure9-1.png)
PDF] Refinements for the amplification and sequencing of red algal DNA barcode and RedToL phylogenetic markers: a summary of current primers, profiles and strategies | Semantic Scholar
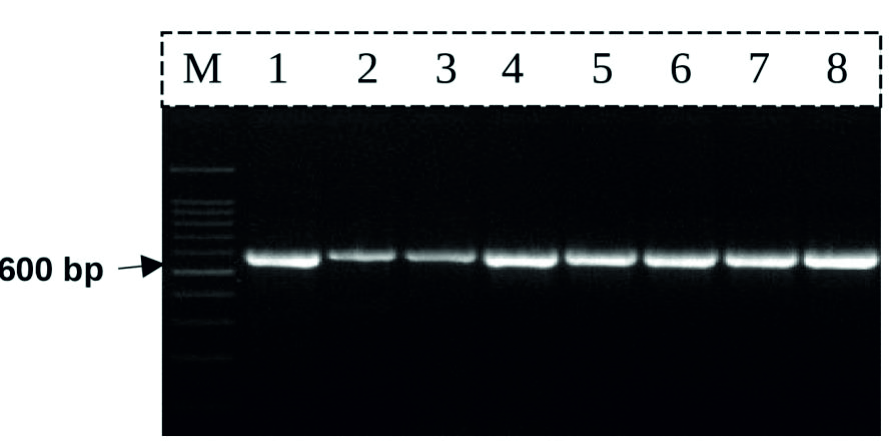
DNA-barcoding and Species Identification for some Saudi Arabia Seaweeds using rbcL Gene - Journal of Pure and Applied Microbiology

DNA barcodes in Egyptian olive cultivars (Olea europaea L.) using the rbcL and matK coding sequences | Journal of Crop Science and Biotechnology

Efficiency of matK, rbcL, trnH-psbA, and trnL-F (cpDNA) to Molecularly Authenticate Philippine Ethnomedicinal Apocynaceae Through DNA Barcoding. - Abstract - Europe PMC
Use of rbcL and trnL-F as a Two-Locus DNA Barcode for Identification of NW-European Ferns: An Ecological Perspective | PLOS ONE

PDF) Use of rbcL and trnL-F as a two-locus DNA barcode for identification of NW-European ferns: an ecological perspective | Arjen de Groot - Academia.edu

Table 2 from Comparative Evaluation of PCR Success with Universal Primers of Maturase K (matK) and Ribulose-1, 5-Bisphosphate Carboxylase Oxygenase Large Subunit (rbcL) for Barcoding of Some Arid Plants | Semantic Scholar

Multiple and different genomic rearrangements of the rbcL gene are present in the parasitic orchid Neottia nidus-avis
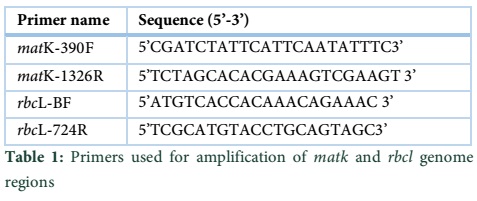
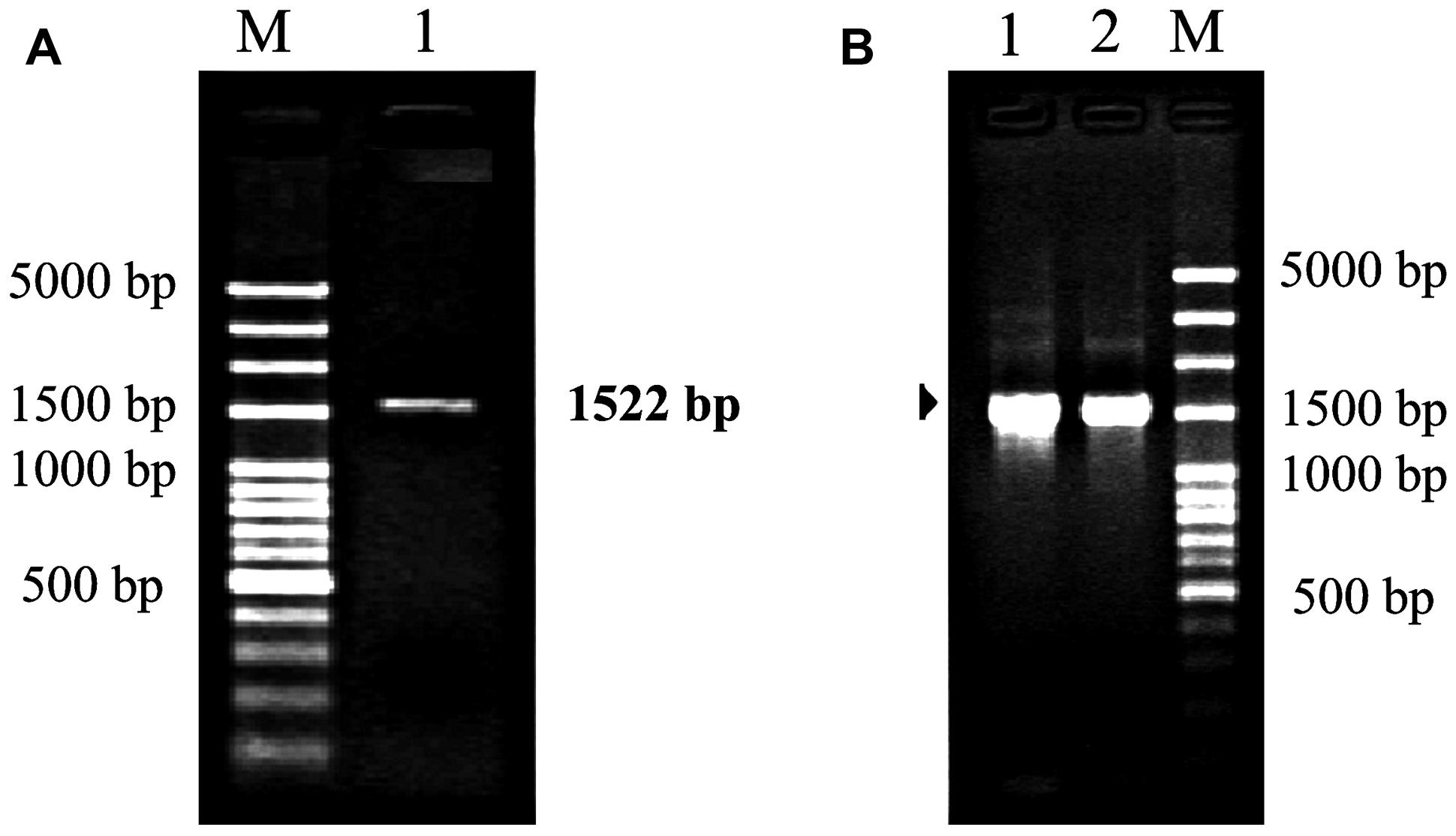
DNA Barcoding: Amplification and sequence analysis of rbcl and matK genome regions in three divergent plant species » Advancements in Life Sciences
Refinements for the amplification and sequencing of red algal DNA barcode and RedToL phylogenetic markers: a summary of current

The design of primer pairs. a Primer pairs positions and lengths of PCR... | Download Scientific Diagram
Amplification and Analysis of Rbcl Gene (Ribulose-1,5-Bisphosphate Carboxylase) of Clove in Ternate Island

Primer design for the chloroplast encoded RuBisCo (rbcL) gene region... | Download Scientific Diagram
Frontiers | Identification of Rubisco rbcL and rbcS in Camellia oleifera and their potential as molecular markers for selection of high tea oil cultivars

Gene elements that affect the longevity of rbcL sequence-containing transcripts in Chlamydomonas reinhardtii chloroplasts | PNAS